totalvi|scvi : Tuguegarao totalVI is a scalable, probabilistic framework for end-to-end analysis of paired transcriptome and protein measurements in single cells. Like other multi-omics analysis methods [ 31, .
Not sure if Its allowed to post this here, but: For the past day and a half, the app just won’t open on the first attempt. I was having issues on desktop as well, but not as bad. I usually have to try to open the app like 4-5 times for it to work and even then usually it doesn’t. I just wait like half an hour then it magically will work again.
PH0 · totalVI — scvi
PH1 · totalVI – streets lab
PH2 · scvi
PH3 · TotalVI: A transformative algorithm
PH4 · Joint probabilistic modeling of single
PH5 · CITE
PH6 · A multi
PH7 · 3. totalVI Tutorial — scVI 0.6.8 documentation
Tons of free Pinay Kabit porn videos and XXX movies are waiting for you on Redtube. Find the best Pinay Kabit videos right here and discover why our sex tube is visited by millions of porn lovers daily. . PINAY KABET KANTOT SA KUSINA HABANG NAGHUHUGAS NG PLATO,HOT FUCK IN THE KITCHEN-PINAY VIRAL . 42,948 .
totalvi*******totalVI is a scalable, probabilistic framework for end-to-end analysis of paired transcriptome and protein measurements in single cells. As with other multi-omics analysis methods .
totalVI is a Python class that uses variational inference to learn a flexible generative model of CITE-seq RNA and protein data. It can be used for dimensionality reduction, .scvi-tools is a Python library that provides various probabilistic models for single-cell omics data analysis, such as scVI, scANVI, Stereoscope, and totalVI. It uses PyTorch, .
totalVI Tutorial¶ totalVI is an end-to-end framework for CITE-seq data. With totalVI, we can produce a joint latent representation of cells, denoised data for both protein and RNA, .CITE-seq analysis with totalVI# With totalVI, we can produce a joint latent representation of cells, denoised data for both protein and RNA, integrate datasets, and compute .
totalVI is a scalable, probabilistic framework for end-to-end analysis of paired transcriptome and protein measurements in single cells. Like other multi-omics analysis methods [ 31, .TotalVI: A transformative algorithm. UC Berkeley researchers have invented a computer algorithm that uses deep learning to integrate gene and protein data about single cells. .totalVI is a Python class that uses neural networks to model the joint distribution of RNA and protein expression in single-cell data. It can be used for various downstream tasks such .totalvitotalVI is a Python class that uses neural networks to model the joint distribution of RNA and protein expression in single-cell data. It can be used for various downstream tasks such .
totalVI is a tool for end-to-end analysis of CITE-seq data, a technique for simultaneous measurement of RNA and protein in single cells. It uses neural networks and Bayesian .
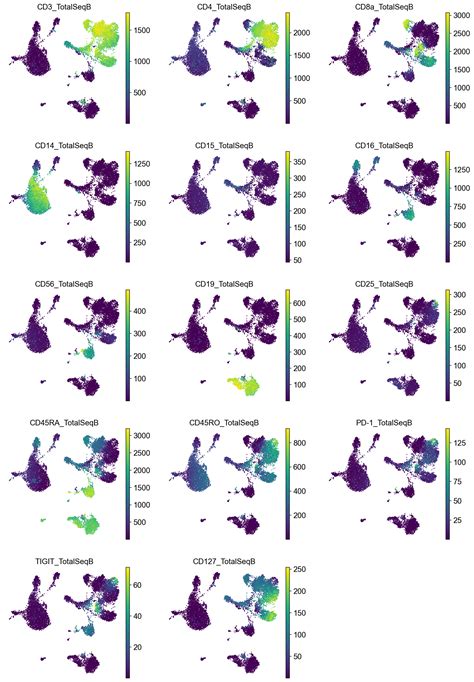
In totalVI, the spike in CD169 is observed for cDC2, but it appears to be small in CD14 monocytes and nearly non-existent in CD16 monocytes. In Seurat 4, the spike in CD169 .
For totalVI, we will treat two CITE-seq PBMC datasets from 10X Genomics as the reference. These datasets were already filtered for outliers like doublets, as described in the totalVI manuscript. There are 14 proteins . 总的来看, TotalVI 和 scVI 功能总的来说是一脉相承的,基于隐变量的几个功能他都有, 基于后验分布的功能它也有,但是由于 TotalVI 多了一个蛋白水平的建模,所以多了两个其特有的protein .totalVI 1 (total Variational Inference; Python class {class} ~scvi.model.TOTALVI) posits a flexible generative model of CITE-seq RNA and protein data that can subsequently be used for many common downstream tasks. The advantages of totalVI are: Comprehensive in . One of the key modeling decisions in MultiVI, compared with previous approaches such TotalVI (ref. 16), is the creation of latent representations for each data modality, followed by the .
Each model (e.g., scVI, scANVI, Stereoscope, totalVI) follows the same user interface that couples nicely with Scanpy, Seurat, or Bioconductor workflows. No more searching through source code. Rapid development of new models .totalVI [1] is a computational method designed for end-to-end analysis of CITE-seq data [2], and developed by Adam Gayoso and Zoë Steier. totalVI relies on neural networks and Bayesian methods to model the observed count data as a composition of biological and technical factors. A major focus of totalVI is to learn a low-dimensional .totalvi scviBSD-3-Clause license. scvi-tools (single-cell variational inference tools) is a package for probabilistic modeling and analysis of single-cell omics data, built on top of PyTorch and AnnData. scvi-tools is part of the scverse project ( website , governance) and is fiscally sponsored by NumFOCUS. Please consider making a tax-deductible donation .
通过这种设计,TotalVI可以从CITE-seq数据中学习配对ADT和mRNA测量的联合概率表示,这些数据解释了每种模态的不同信息。 同样,对于SNARE-seq或SMAGE-seq数据,Cobolt和scMM采用多模态变分自编码器对多种模态进行联合建模,并学习单细胞mRNA-seq和ATAC-seq数据的联合嵌入。

Documentation#. scvi-tools (single-cell variational inference tools) is a package for end-to-end analysis of single-cell omics data primarily developed and maintained by the Yosef Lab at UC Berkeley and the Weizmann Institute of Science. scvi-tools has two components:. Interface for easy use of a range of probabilistic models for single-cell omics (e.g., scVI, .To evaluate totalVI's performance, we profiled immune cells from murine spleen and lymph nodes with CITE-seq, measuring over 100 surface proteins. We demonstrate that totalVI provides a cohesive solution for common analysis tasks such as dimensionality reduction, the integration of datasets with different measured proteins, estimation of . To evaluate totalVI’s performance, we profiled immune cells from murine spleen and lymph nodes with CITE-seq, measuring over 100 surface proteins. We demonstrate that totalVI provides a cohesive solution for common analysis tasks such as dimensionality reduction, the integration of datasets with different measured proteins, .
totalVI reproducibility This repository contains the code to reproduce the analyses performed in the manuscript "Joint probabilistic modeling of paired transcriptome and proteome measurements in single cells".
scviIncluded with Total Studio 4 MAX is the new IK Product Manager, which automates the installation process and keeps all the IK products organized and up to date. Now you can focus on making music instead of updating .
之前听师兄在组会上介绍了scvi,恰巧最近在用scvi做一些分析,感觉这个方法很精巧效果也不错,因此研究了一下原始论文,这里简要介绍一下这个单细胞数据分析工具的新星。. scvi 是Nature Methods在2018年12月发表的一个单细胞RNA测序 (scRNA-seq)数据的集成分析平 .
The totalVI model contains two different components that can be used for downstream analysis. First, totalVI contains a low-dimensional representation of cells, known as the latent space, which contains information from both the RNA and protein data while controlling for noise and technical artifacts in each data type. TotalVI is a deep variational autoencoder that can capture the same latent space of different data types 25. With this design, TotalVI can learn a joint probabilistic representation of the paired . totalVI identifies differentially expressed genes and proteins. totalVI intersect was applied to the SLN-all dataset. a, UMAP plot of SLN-all, after clustering and annotating the data (Methods 4.11). b, c, Heatmap of markers derived from one-vs-all tests for (b) RNA and (c) proteins. For each cell type, we display the top three protein markers . Supplementary Note 3 presents a case study of applying this approach with totalVI by projecting data from patients with COVID-19 into an atlas of immune cells.
Ethereum casinos are online gambling sites that accept the cryptocurrency Ethereum as a payment option, providing an easy and secure way to make wagers. How We Rate Casinos. We evaluate casinos based on four primary criteria to identify the best options for US players. We ensure that our recommended casinos maintain high standards, giving .
totalvi|scvi